Supporting Early Career Researchers
In collaboration with the Board of Directors, the GI Research Foundation’s Associates Board focuses on funding cutting-edge research by supporting diverse physicians and scientists early in their careers.
2023 Young Investigator Awards
Each year, the GI Research Foundation’s Associates Board awards early career grants to young investigators, providing a springboard at a critical time in an investigator’s scientific career. In the 2023 grant cycle, the Associates Board awarded $40,000 to four early career investigators at the University of Chicago in support of their novel research projects.
The Projects
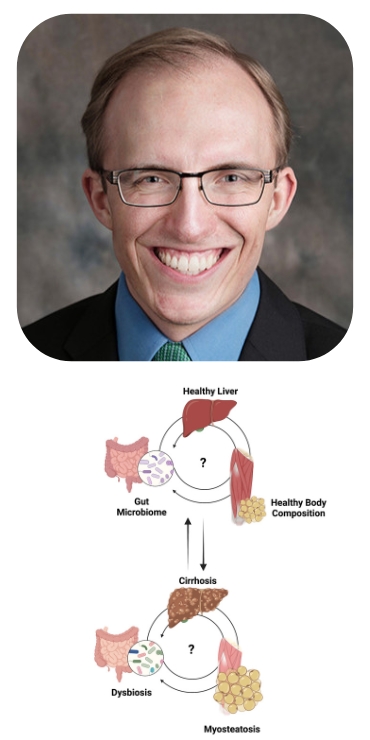
Interaction of body composition and the microbiome in liver disease
Researcher: Alan Hutchinson, MD, PhD
Award: $10,000
The University of Chicago Digestive Diseases Center leads the nation in developing approaches to understand and fight disruptions in healthy nutrition and to understand the role of the bacterial community in the gut – the gut microbiome – in those disruptions. Malnutrition leads to changes in muscle and fat quality, quantity, and location. Muscle and fat are metabolically active organs, with metabolites that communicate with and are modified by the gut microbiome and liver. How these networks are disrupted by and exacerbate liver disease is poorly understood.
The Duchossois Family Institute (DFI) has collected and analyzed the metagenomic (i.e. microbial composition) and targeted metabolomic (i.e. functional microbial products) of over 450 patients with liver disease hospitalized at the University of Chicago. This dataset provides a unique opportunity to make gain insight into the interplay of the microbiome and liver disease.
Muscle wasting (i.e. sarcopenia), fat infiltration of muscle (i.e. myosteatosis), and fat location (i.e. visceral vs subcutaneous adiposity) can be identified from CT scans using imaging tools. Over 300 patients have CT scans collected within days of their DFI stool and serum collection, many with repeats across multiple hospitalizations. We propose to determine how the body composition of these patients interacts with their gut microbiome and liver disease.
Aim 1: To determine the effect of body composition on predicting adverse outcomes and overall mortality in hospitalized patients with cirrhosis.
Aim 2: To determine the association of changes in body composition with changes in the gut microbiome over time.
Role of Akkermansia muciniphila in mucosal healing—a potential microbial therapeutic approach to IBD
Researcher: Deepinder Kaur, PhD
Award: $10,000
Akkermansia muciniphila has been shown to be an abundant bacterial species in mice and humans. Its abundance is decreased in IBD patients and patients with metabolic disease. A. muciniphila adheres to intestinal epithelial cells and increases barrier integrity, which is critical to the prevention of penetration of the epithelial layer by microbes.
In the Chang lab, we observed that the expression of Hsp25 in the intestine is crucial for wound healing in a colitis mouse model. After colitis induction, transgenic mice that overexpress Hsp25 only in the intestinal epithelial compartment demonstrate an increased abundance of A. muciniphila, which is associated with accelerated healing. Therefore, we hypothesize that A. muciniphila promotes wound healing by producing specialized factors that induce intestinal epithelial cell proliferation.
To this end, we will examine:
Aim 1: Whether heat-killed A. muciniphila leads to wound healing.
Aim 2: Factors of A. muciniphila that interact with host.
The available biologic therapies today focus on suppressing the immune mechanisms that destroy the epithelium and deplete the mucus layer. Furthermore, many immunomodulatory therapies induce remission in only a minority of patients. Thus, a new generation of therapies can be developed to target epithelial repair directly and can complement the immunetargeting therapies currently available. Studying the microbial factors and pathways by which they induce epithelial repair can result in more efficacious therapeutic treatments for IBD.
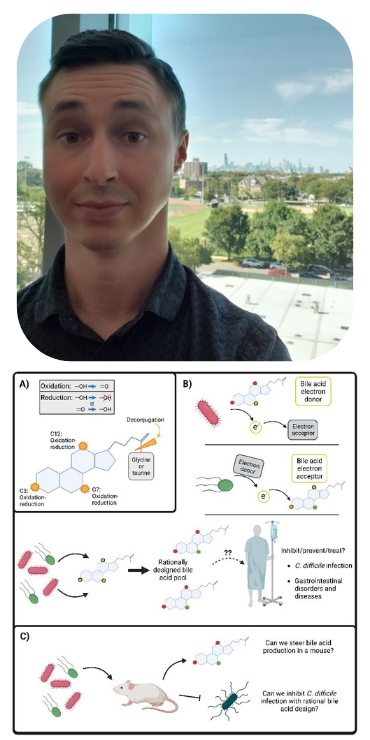
Bacterial anaerobic respiration and rational bile acid design
Researcher: Alexander Little, PhD
Award: $10,000
Bile acids were initially only considered to be molecules with detergent properties used for the uptake of fats and fat-soluble vitamins in the intestines. They are now known to be complex digestive hormones regulating both host physiology and bacterial competition. Within the anaerobic (oxygen-depleted) gut environment, bacteria compete for not only available nutrients but also sources of generating energy. While the majority of microbes perform fermentation for generating energy, a certain subset of microbes possess a diverse array of reductase enzymes in order to utilize molecules other than oxygen to respire with (essentially the moving of electrons) and generate energy. Bile acids are known to be modified from their initial excreted ‘primary’ form to over 50 chemically distinct ‘secondary’ bile acids by bacteria within the intestines, the purpose of which largely remains unknown.
The following aims center around connecting complex bile acid modification to anaerobic respiration, determining if bile acid pools can be pushed in planned directions, and ultimately what that means in the interplay between the microbiome and human physiology.
Aim 1: Determine if bacteria with diverse respiratory capabilities within the microbiome (Eggerthellaceae, Burkholderiaceae, Erysipelotrichaceae) utilize different bile acids as either electron donors or electron acceptors in anaerobic respiratory chemistry.
Aim 2: Investigate if bile acid composition or usage can be steered with certain combinations of electron donors or electron acceptors. If time and finances permit, we will attempt to colonize mice with rationally designed groups of bacteria to determine if can modify bile acid production in vivo. In the long term, this may inform on rational probiotic and/or diet design to drive favorable bile acid pool compositions.
We believe that with further understanding of bacterial usage of bile acids in anaerobic respiration, we could predictably couple certain bacteria and substrates to drive the bile acid pool into particular reduced or oxidized forms. This rational design could be used for the benefit of human health.
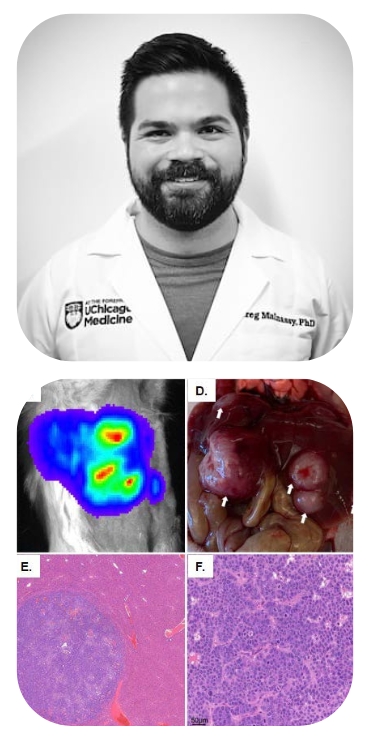
Targeting the unfolded protein response to enhance PanNETs' radiosensitivity
Researcher: Gregory Malnassy, PhD
Award: $10,000
Since its FDA approval in 2018, peptide receptor radionuclide therapy (PRRT) has represented a paradigm shift in the treatment of advanced and/or metastatic pancreatic neuroendocrine tumors (PanNETs), given its ability to deliver targeted radiation directly to tumor cells. Despite its increasingly widespread use, however, PRRT serves only as a palliative as opposed to curative modality, as the majority of patients will experience disease progression due to either intrinsic or acquired therapy resistance. Furthermore, higher doses of PRRT place patients at increased risk of radiation toxicity. Consequently, novel adjuvants are desperately needed that will not only enhance the efficacy of PRRT but also mitigate toxicity to nontarget organs. We have evidence to support the hypothesis that PanNETs activate an ancient stress adaptation pathway called the Unfolded Protein Response (UPR) to resist radiation-induced cell death, and that small molecule inhibitors of the UPR enhance PanNETs’ radiosensitivity.
We will test this hypothesis via two aims:
Aim 1: Determine how UPR induction promotes radioresistance in PanNETs in vitro.
Aim 2: Determine whether the UPR can be targeted to potentiate radiation in murine PanNET models.
Our research project will provide powerful mechanistic insights into the role of the UPR on PanNET radiotherapy response and use of UPR inhibitors to improve PRRT. If successful, our results could quickly translate into a clinical trial of this combination in humans.

